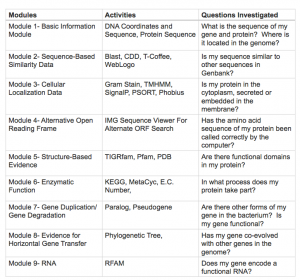
The GENI-ACT is a package of nine independent modules that will allow students and teachers to annotate a gene from a bacterium. Annotation is the process of assigning biologic function to to a gene. The 9 modules of GENI-ACT and their purpose are shown in the figure below:
-
-
- Use and master multiple database analysis software packages related to bioinformatics.
- Strengthen library and web-search skills
- Develop skills in making hypotheses and the design of experiments to test them
- Sharpen skills in analysis, synthesis and in the presentation and interpretation of results
- Experience the collaborative nature of science.
-
See how gene annotation demonstrates the use of the scientific method.
The project will target teacher populations as follows:
The project is broken down into three main components:
Summer Teacher Training Workshop:
A summer training workshop for teachers who will subsequently guide their students to perform basic gene annotations later in the project will take place in July. Teachers will have a clinical focus to their training, by (i) receiving training in biomedical applications of bioinformatics during their summer training, and (ii) focusing on genes of clinical significance in human microbial pathogens (e.g. virulence genes, antibiotic resistance genes). The significance of annotation to novel gene discovery, to anchoring of metagenomic sequencing data and to the accuracy of gene calling from such a unique organism will be stressed. Experts in clinical bioinformatics or molecular diagnostic fields will participate in the teacher training workshop. The summer training workshop for teachers will have 5 continuous days of training in one week over the summer. Teachers will then come back to UB for the equivalent of 2 refresher training days in the fall to refresh their gene annotation skills prior to having them work with their students for the semester 2 gene annotation and then for another day in the spring to help them with the review of their students’ progress and to help them prepare for the final capstone poster presentations. Teachers still have a total of 8 days of training in the project and be paid for their time in training and in working with their students.
Semester 1 Activities:
The Semester 1 activities for the SEPA project will build student knowledge in the areas of molecular diagnostics and clinical genomics. We envision 5 activity modules to achieve this goal:
- Activity 1: “Spotlight on Health Professionals and Genomics” including videos of interviews with those individuals discussing how and why they entered the field. Students will be offered career guidance by AHEC facilitators at the end of this activity as well.
- Activity 2: “Building a Company Role Play” as a group, students will explore the different types of positions necessary to build a successful bioscience company. This activity is meant to expose students to the variety of career opportunities in STEM fields.
- Activity 3: SEPA Project Overview Presentation by Dr. Stephen Koury or Dr. Rama Dey-Rao is designed to introduce your students to the work they would be completing during the school year if selected as part of the treatment group. Dr. Koury and Dr. Dey-Rao review the nine modules in GENI-ACT and answer student questions.
Semester 2 Gene Annotation:
The project will involve gene annotation of prokaryotes. The organization of genes is simpler in prokaryotes and the complications of alternative splicing are avoided. Annotations of prokaryotic genes are therefore likely to be more easily grasped by high school level students.
The project will use genomes of multiple clinically significant microorganisms in Genbank as subjects for the annotation activities. These will be chosen to complement discussions in Semester 1 and will involve students doing research to generate background information about the organism, the types of infection it causes, the outcomes that result from infection and whether molecular diagnostic tests exist for their organism. Each school will be assigned a different organism on which to work. A mixture of genes will be selected from each organism for the students to annotate.
Examples of work on the genome annotation project performed by high school students and MS level students at the university at Buffalo can be seen in the Student Research page of this website.
Interested in participating?
Please see the official program recruitment flyer and contact the coordinator of the project in the AHEC representing your school district on the Contact Information Page.
